 |
The Analyze - Composition and Tm windows display the nucleotide composition and melting temperatures of the selected Forward or Reverse Primer, Upper or Lower Probe, Current Oligo, entire sequence, or PCR product. Various calculation methods are used for comparison.
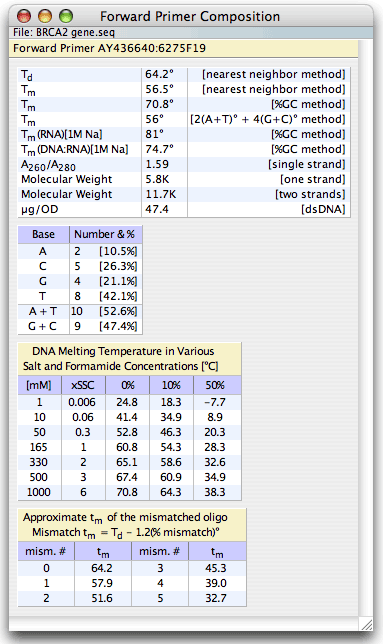
The expression “Td” is the melting temperature calculated by the nearest neighbor method for 100 nM oligos in 1M monovalent salt concentration between a given oligo and its complement, so that the dangling ends and mismatches don’t count. On the other hand, the "Tm for the nearest neighbor method" is shown for the nucleic acid at salt conditions set in the Change – Search Parameters window. It is a ‘full’ calculation, that includes the dangling ends, however the mismatches, if present, are not considered.
Besides the Tm values the Composition and Tm windows display:
• The A260/A280 absorption ratio
• The molecular weight of oligo, both single stranded and double stranded
• The number and percentage of each base in the oligo plus the number and percentages of A+T and of G+C in the sequence
• The melting temperature (%GC method) of the selected nucleic acid molecules with their complements at various salt concentrations (expressed both as mM and as multiples of the SSC buffer) in 0%, 10%, and 50% formamide
• Calculation of melting temperatures (nearest neighbor method) for the short oligos, with up to five non-specified mismatches.
|
![]()
![]()
